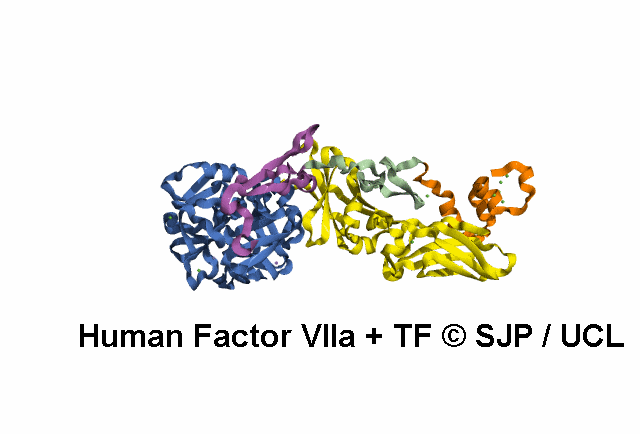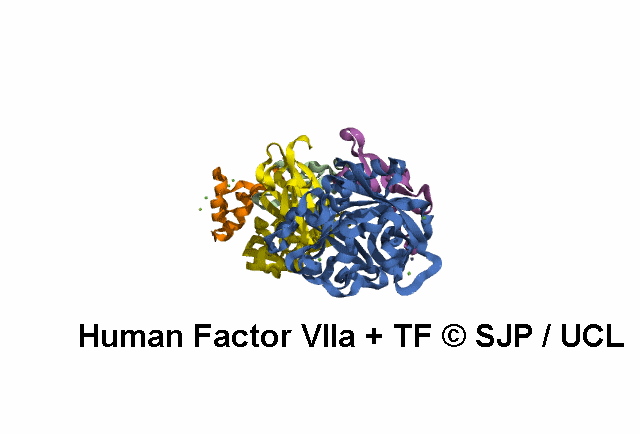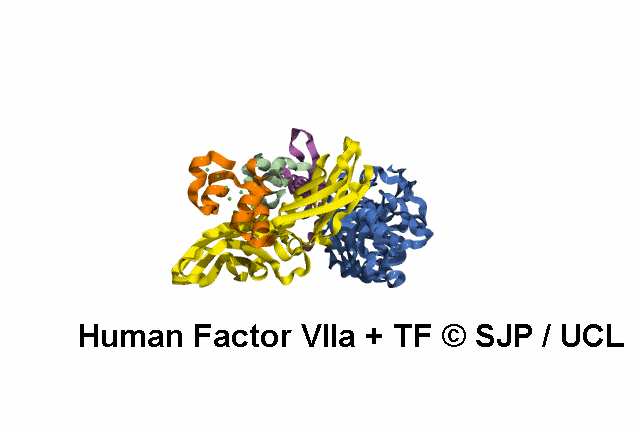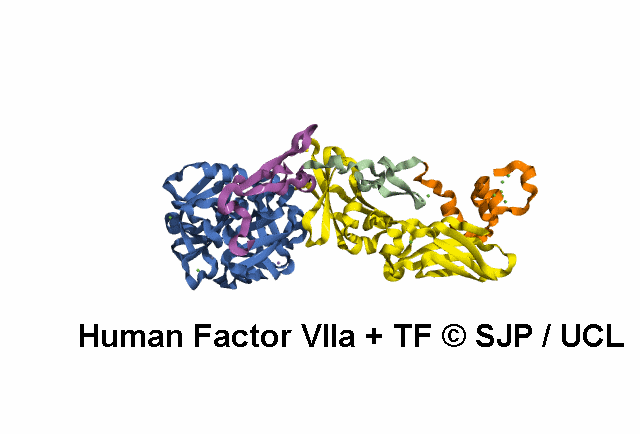
In Depth Variant Analysis: c.1268G>T (p.Ser423Ile)
c.1268G>T
p.Ser423Ile (Legacy AA No. 363)
Mutation Type:
Point
Domain:
Serine Protease
Codon Change:
AGC > ATC
Mutation Effect:
Missense
Location:
Exon(9)
No of bases:
1
No. of patients reported:
3
Patient Information : Show
Residue Information :
| Name | Type | Cyclic | Size | Hydrophobicity | Charge | |
|---|---|---|---|---|---|---|
| |
|
|
|
|
|
|
| |
|
|
|
|
|
|
Substitution Analysis :
Structural Implications :
Hint: Left click mouse | To rotate the structure
Hint: Rotate mousewheel | To zoom in/out the structure
Hint: Right click mouse | to use applet control options
|
Display Options
Right Click on the molecule's screen for more options.
NOTE: If a Java error message appears, please click the following LINK.